NxSeqЂч 40 kb Mate-Pair Cloning Kit
True long-span, mate-pair libraries for Next Generation Sequencing
- Complete your genome with 40 kb, long-span, mate-pair NGS libraries
- Skip the adapters: pNGS vector contains built-in Illumina and Roche 454 primers sites
- Ideal for de novo, whole genome sequencing projects
- Transcription free vector: highly stable clones
Introducing a new paradigm in long-span, mate-pair Next Gen libraries:
the NxSeq 40kb Mate-Pair Cloning Kit.
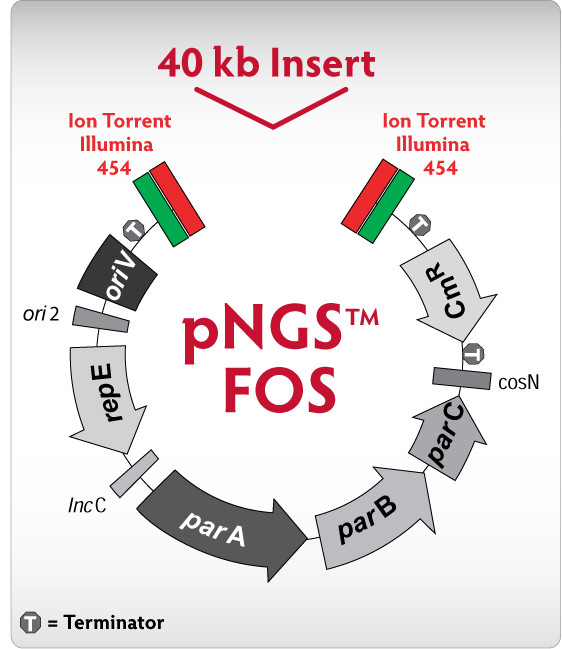
The pNGS FOS vector (fig. 1) represents the combination of Lucigen's patented transcription free vectors with a first-of-its-kind feature: built-in Roche 454 and Illumina primer binding sites – no adapter ligation necessary! Using a 40 kb paired-end library, you can now build the critical scaffold that enables a truly complete genome assembly. Get exceptionally stable clones due to our transcription-free vector, reduce time and uncertainty by skipping adapter ligation, and get more sequence coverage with less bias than ever before.
Unique Workflow:
Let Lucigen make the process of generating long-span, mate-pair libraries easier than ever before.
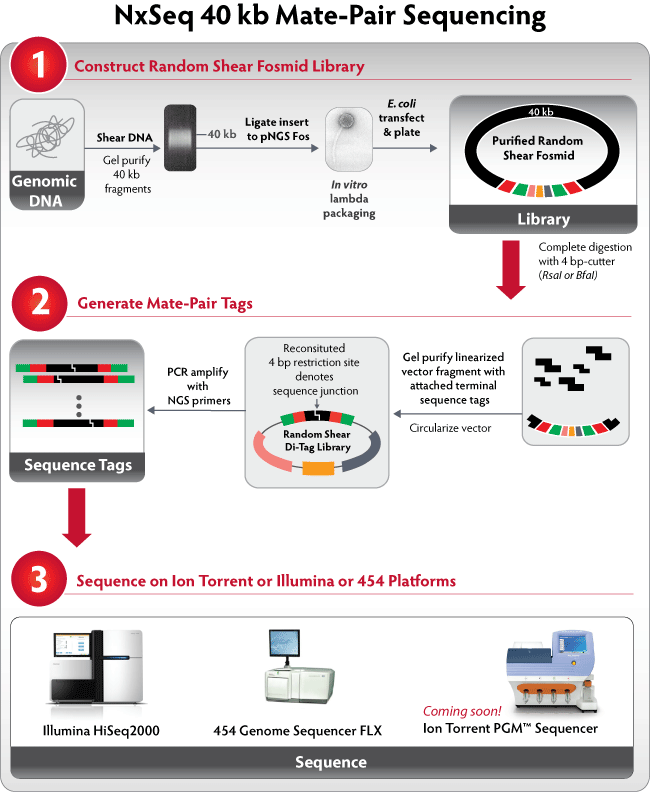
- Shear and size select for 40 kb fragments, ligate to the pNGS FOS vector and perform lambda packaging.
- Cut with a 4bp restriction enzyme (RsaI, CviQI, BfaI, or FspBI)
- Reconstitute the restriction site to create a known mate-pair junction.
- Amplify with either Roche 454 or Illumina primers and proceed to sequencing (fig. 2).
Figure 2.
High Efficiency 40 kb Paired-end Sequencing for Next Generation Platforms
To demonstrate the efficiency of cloning and sequencing using the pNGS FOS vector, large-scale, long-span, mate-pair sequencing of a human cell line (GM15510, Coriell) using Illumina technology resulted in 64% of filtered reads accurately mapping to the genome (Table 1). This represents many-fold higher efficiency than existing systems and will allow the accurate assembly of genomes for the first time using next gen sequencing platforms.
Table 1. Summary of Illumina long-span, mate-pair sequencing of a human cell line |
| Stage |
No. of Reads |
% |
| Pipeline |
8,683,854 |
ЁЁ |
| Filtered reads |
6,506,126 |
ЁЁ |
| Reads mapped |
5,173,778 |
79.2 |
| Reads with pair-sequences |
4,685,996 |
90.6 |
| Long-span, mate pairs* |
3,018,576 |
64.4 |
| *Mapped on same chromosome, ~40 kb apart, uniquely mapped.
|
 IDT
IDT