* 2019 РЬШФ УыБо ОШЧд*
PyroPhageЂч 3173 DNA Polymerase, Wild Type
- Strong proofreading activity.
- Can be thermocycled for PCR.
Superior DNA amplification with the first thermostable phage DNA polymerase
LucigenЁЏs PyroPhage 3173 DNA Polymerase (patent pending) is the first thermostable enzyme to offer the superior DNA replication performance of a phage replicase. Because all current PCR enzymes are derived from bacterial or archaeal DNA repair polymerases rather than true DNA replicases, they have inherent limitations in DNA replication fidelity, robustness, and processivity. PyroPhage 3173 DNA Polymerase, the first replicase cloned from bacteriophage inhabiting a boiling hot spring (Figure 1), avoids these limitations for much more effective DNA amplification.
The thermostability of PyroPhage 3173 DNA Polymerase makes it useful for thermocycling methods, including PCR. The potent 3ЁЏ-5ЁЏ exonuclease (proofreading) activity of the Wild Type enzyme results in extraordinarily high fidelity.
PyroPhage 3173 DNA Polymerase Advantages
Superior PCR amplification
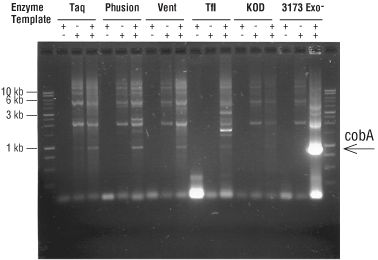
As shown in Figure 2, PyroPhage 3173 DNA Polymerase is much more effective than current PCR enzymes in amplifying certain difficult templates. PyroPhage 3173 DNA Polymerase effectively amplifies most templates of up to 4 kb. Because of the strong proofreading activity of the wild type enzyme, it is highly recommended to use phosphorothioated primers.
Figure 2. PCR amplification of a difficult template. PCR amplification of the Bacillus cobA gene (GC rich; strong secondary structure) using PyroPhage 3173 (Exo Minus) or the indicated DNA polymerase.
Superior replication fidelity.
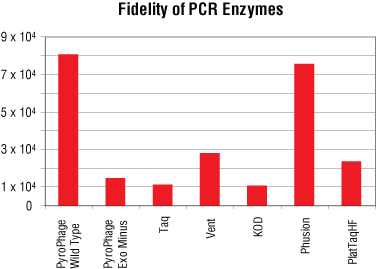
PyroPhage 3173 DNA Polymerase (Wild Type) demonstrates extremely strong proofreading activity and copy fidelity (Figure 3). Because of this very active nuclease activity, use of phosphorothioated primers is recommended when using the Wild Type enzyme for DNA amplification.
Figure 3. PyroPhage 3173 fidelity. Fidelity measurements, shown as the ratio of correct to incorrect nucleotide incorporations, are based on the LacIq forward mutation assay (1, 2). PyroPhage 3173 DNA Polymerase (Wild Type and Exo Minus) was compared to various commercially available enzymes.
Superior amplification efficiency.
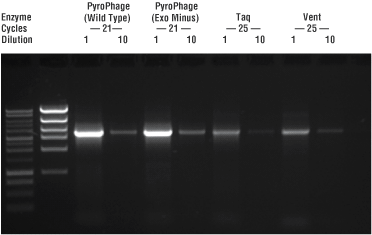
PCR with PyroPhage 3173 DNA Polymerase results in higher product yields in fewer cycles (Figure 4).
Figure 4. PyroPhage 3173 amplification efficiency. The PCR amplification efficiency of PyroPhage 3173 DNA Polymerase (Wild Type & Exo Minus) was compared to those of Taq and VentЂч DNA polymerases using the fidelity assay template from Figure 2. Reaction products formed after the indicated number of cycles were quantified by densitometry of the gel bands. Cycle numbers for the PCRs are shown. Efficiency was calculated by published methods (1).
Convenient and easy to use.
PyroPhage 3173 DNA Polymerase comes with PyroPhage 3173 2X PCR Buffer containing the magnesium concentration optimal for most PCR applications, PCR Control + Primers, and complete protocols.
PyroPhage 3173 DNA Polymerase Properties
Table 1. Established characteristics of PyroPhage 3173 DNA Polymerase |
|
PyroPhage 3173 DNA Polymerase |
Wild Type |
|
3ЁЧ-5ЁЧ exonuclease |
strong |
|
5ЁЧ-3ЁЧ exonuclease |
none |
|
Extension from nicks |
strong |
|
Thermostability (TЈі @95ЁЦ in PCR buffer) |
10 min. |
|
Km dNTPs |
40 µM |
|
Km DNA |
5.3 nM |
|
Processivity |
not determined |
|
Fidelity |
8 ЁП 104 |
|
3ЁЧ ends of amplicons |
blunt |
Usage Information
Certain primer/template sets are more effectively amplified by PyroPhage 3173 DNA Polymerase. Amplification of other apparently similar primer/template sets can be problematic. The basis of this inconsistency is not understood. Because of this variability, no predictions can be made on the effectiveness of PyroPhage 3173 DNA Polymerase in amplifying a given primer/template set. Variability may be significantly reduced by use of alternative primer sets or common PCR additives, such as betaine or ectoine.
Legal Information
The PyroPhage DNA Polymerases and methods of using the PyroPhage DNA Polymerases are covered by pending patents assigned to Lucigen Corporation. Lucigen does not encourage or support the unauthorized or unlicensed use of the PCR process, reverse transcription PCR process or isothermal DNA synthesis. It is the sole responsibility of the buyer to ensure that use of the product does not infringe the patent rights of third parties.
PyroPhage 3173 2X PCR Buffer: 40 mM Tris-HCl, 20 mM (NH4)2SO4, 20 mM KCl, 4 mM MgSO4, 0.2% Triton X-100, thermoprotectant, pH 8.8 @ 25ЁЦC.
Activity Assay
Polymerase activity is assayed at 70ЁЦC with 0.2 mM each of dATP, dGTP, dTTP, dCTP (mix of unlabeled and [33P] dCTP); 10 µg activated calf thymus DNA, and 0.1 mg/ml BSA in a final volume of 50 µl.
Absence of Endonuclease
PyroPhage 3173 DNA Polymerase, WT is determined to be free of detectable endonuclease or nicking activity. Supercoiled plasmid DNA is incubated with enzyme for 16 hours at 70ЁЦC. Agarose gel electrophoresis shows no alteration in mobility consistent with endonuclease or nicking activity.
Absence of Exonuclease
PyroPhage 3173 DNA Polymerase, WT is tested to be free of contaminating exonuclease activity by incubating Hind III-digested lambda DNA with enzyme at 70ЁЦC for 16 hours. Agarose gel electrophoresis shows no alteration in mobility consistent with exonuclease activity.
Absence of Ribonuclease
PyroPhage 3173 DNA Polymerase, WT is tested to be free of contaminating RNAse activity by incubating with a fluorogenic RNAse substrate for 1 hour at 37ЁЦC. No increase in assay fluorescence above background was detected.
Functional Assay
PyroPhage 3173 DNA Polymerase, WT is tested for performance in PCR using the PyroPhage 3173 PCR Buffer to amplify a 2 kb region of the endA gene from 2 X 106 molecules (10 ng) of E. coli gDNA. The resulting PCR product is visualized on an ethidium bromide-stained agarose gel.
 IDT
IDT