*2019Гт РЬШФ УыБо ЧЯСі ОЪРН
Bst DNA Polymerase, Exonuclease Minus
- Highest strand displacement activity available.
- Used in isothermal amplification and Next Generation Sequencing.
High specific activity for DNA amplification and sequencing.
Applications
- Strand displacement amplification*
- DNA sequencing through high GC regions (1,2)
- Rapid sequencing from nanogram amounts of DNA template (3)
Bst DNA Polymerase, Exonuclease Minus, is a recombinant form of the 67 kDa Bacillus stearothermophilus DNA Polymerase protein (large fragment). The enzyme has 5ЁЏ-3ЁЏ polymerase activity and strand displacement activity, but it lacks 3ЁЏ-5ЁЏ exonuclease activity. It also has reverse transcription activity.
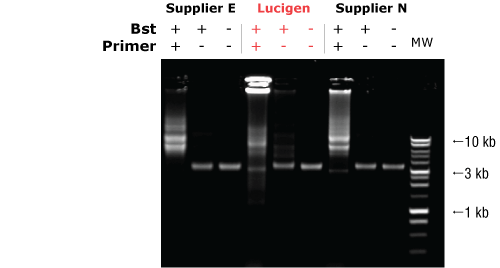
Lucigen's Bst DNA Polymerase, Exonuclease Minus, has higher strand displacement activity than that of other suppliers (Figure 1). The enzyme can be used in nucleic acid amplification methods* such as isothermal amplification, whole genome amplification (WGA), and multiple displacement amplification (MDA). It also can be used in Next Generation sequencing.
This enzyme has optimal activity at 65ЁЦC. It is suitable for sequencing DNA with high GC content and secondary structures. It is available in concentrations of 8,000 U/ml or 50,000 U/ml.
Bst DNA Polymerase, Exonuclease Minus, is supplied with 10X DNA Polymerase Buffer B, composed of 200 mM Tris-HCl pH 8.8, 100 mM (NH4)2SO4, 100 mM KCl, 20 mM MgSO4, and 1.0 % Triton X-100.
Figure 1. Lucigen Bst DNA Polymerase, Exonuclease Minus, possesses greater strand-displacing polymerase activity. M13 single stranded DNA was incubated with or without 8 units of Bst DNA Polymerase(+/- Bst) in reaction buffer supplied by the manufacturer, with or without replication primer (+/- primer) for 30 minutes at 65ЁЦC. MW, 1 kb ladder.
Tips for use:
- Requires 0.1% Triton X-100 for long term storage.
- Reaction temperatures above 70ЁЦC are not recommended.
- Bst DNA Polymerase cannot be used for thermal cycle sequencing.
Heat Inactivation: 80ЈЌC for 20 min.
References:
- Griffin, H. and Griffin, A. (1994) PCR Technology, 228-229.
- McClary, J. et al. (1991) J. DNA Sequencing and Mapping, 1, 173-180.
- Mead, D.A. et al. (1991) Biotechniques, 11, 76-87.
Storage Buffer: 10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM DTT, 0.1 mM EDTA, 0.1% Triton X-100, and 50% Glycerol.
Purity: >99% pure by SDS-PAGE. No detectable DNA contamination. 10 µl of enzyme at 8 U/µl of the sample was tested for E. coli genomic DNA contamination by PCR amplifying with the E. coli 16S ribosomal primers.
Activity Determination: One unit catalyzes the incorporation of 10 nmol of dNTP into acid-insoluble material in 30 minutes at 65ЁЦC in 20 mM Tris-HCl pH 8.8, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1 % Triton X-100, 30 nM M13mp18 ssDNA, 70 nM M13 sequencing primer(-47) 24 mer 200 µM dGTP, dATP, dTTP, dCTP (a mix of unlabeled and and 33P]dCTP), and 0.1 mg/ml BSA.
Absence of Endonuclease or Nicking Activity Incubation of 8 U and 50 U of Bst DNA Polymerase, Exonuclease Minus, with 1 µg of supercoiled pBR322 DNA for 16 hours at 37ЁЦ and 65ЁЦC resulted in no detectable conversion to relaxed or linear forms by agarose gel electrophoresis.
Absence of Exonuclease Activity: Incubation of 8 U and 50 U of Bst DNA Polymerase, Exonuclease Minus, with 1 µg of HindIII-cut lambda DNA for 16 hours at 37ЁЦ and 65ЁЦC resulted in no smearing of bands on agarose gels. Single stranded and double stranded exonuclease activities were tested by incubating 10 µl of enzyme at 8 U/µl with radiolabeled DNA substrate for one hour at 37ЁЦ and 65ЁЦC, resulting in less than 0.1% release of TCA-soluble counts.
 IDT
IDT